Plant Genomics
Background
Objectives of the Work
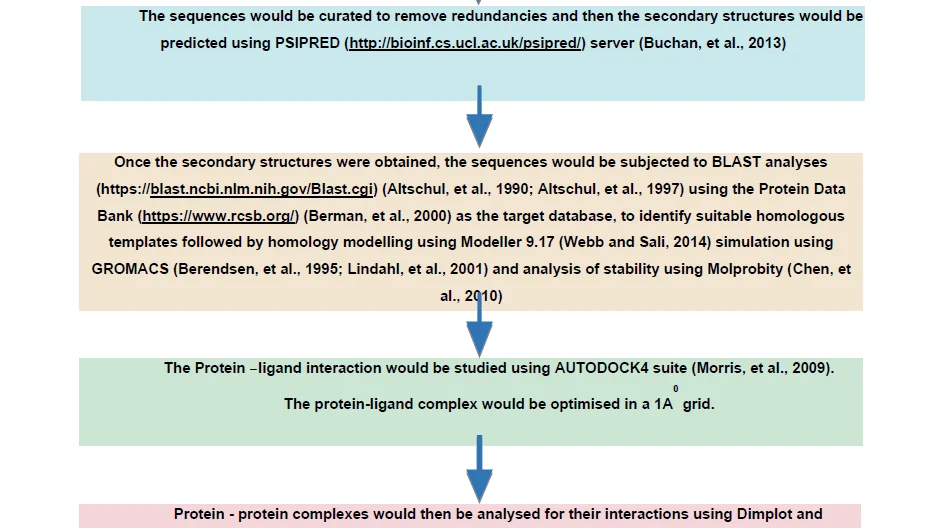
Case Study 1
Lawsonia inermis (Mehendi)
Lawsonia inermis L., commonly known as Henna plant, have been reported to be useful both medicinally and commercially. The phytochemical extract of this plant leaves have been reported to be a rich source of phenols, glycosides, anthraquinones and many other active constituents. Therapeutically, this plant has been used to treat diabetes, arthritis, obesity, ulcers, wounds, microbial infections, inflammation and liver damage. The extracts have also been instrumental in lowering blood sugar and cholesterol levels in mice. But one of the most crucial aspects of this plant is, limiting the growth of malignant cells. Extracts have shown apoptosis promoting activity in human cancer cell lines like breast cancer cells. The pigment lawsone, is commercially used on a large scale, as a dying agent for fabrics and skin. This component has shown potential role in reducing oxidative burst in cell, hence, establishing its role as an antioxidant, which should help researchers to manipulate the property, for establishing new potential drugs against cancer. Apart from “Lawsone” the small molecule reservoir of Lawsonia inermis L. have not been commercially utilized effectively and in the future these bioactive compound set should be explored for formulating new chemical entities.
Vigna mungo transcriptome profiling
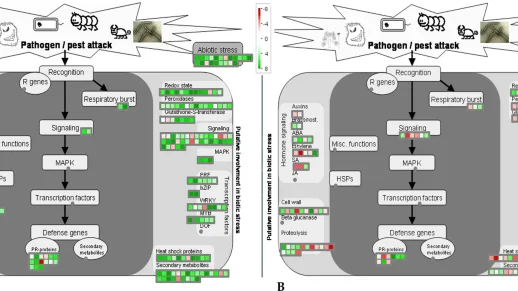
Mungbean Yellow Mosaic India Virus (MYMIV)-infection creates major hindrance in V. mungo cultivation and poses significant threat to other grain legume production. Symptoms associated include severe patho-physiological alterations characterized by chlorotic foliar lesion accompanied by reduced growth. However, dissection of the host’s defense machinery remains a tough challenge due to limited of host’s genomic resources. A comparative RNA-Seq transcriptomes of resistant (VM84) and susceptible (T9) plants was carried out to identify genes potentially involved in V. mungo resistance against MYMIV. Distinct gene expression landscapes were observed in VM84 and T9 with 2158 and 1679 differentially expressed genes (DEGs), respectively. Transcriptomic responses in VM84 reflect a prompt and intense immune reaction demonstrating an efficient pathogen surveillance leading to activation of basal and induced immune responses. Functional analysis of the altered DEGs identified multiple regulatory pathways to be activated or repressed over time. Up-regulation of DEGs including NB-LRR, WRKY33, ankyrin, argonaute and NAC transcription factor revealed an insight on their potential roles in MYMIV-resistance; and qPCR validation shows a propensity of their accumulation in VM84. Analyses of the current RNA-Seq dataset contribute immensely to decipher molecular responses that underlie MYMIV-resistance and will aid in the improvement strategy of V. mungo and other legumes through comparative functional genomics.
Case Study 2
Dr. Anirban Kundu, Assistant Professor, RKMVC, Rahara
Case Study 3
Research Team
- Prof. Amita Pal (Emeritus Scientist, Bose Institute, Kolkata)
- Prof Pratiti Ghosh (Professor, WBSU)
- Dr. Debleena Roy (Lady Brabourne College, Kolkata)
- Dr. Subhadeepa Sengupta (Bidhannagar College, Kolkata)
- Dr. Anirban Kundu (RKMVC, Rahara)
- Dr. Pankaj K Singh ( Western Sydney University)
- Dr. Sayak Ganguli (SXC_Kolkata)
Interns
- Saptaki De (BMBT – SXC, Kolkata)
- Saptarshi Bhattacharya (BMBT – SXC, Kolkata)
- Mahima Chaudhury (VIT, Vellore)
Project Duration
- Open ended
- Funding: PUPA and SXC Intramural; Several Completed Projects under State and Central Funding Agencies.
